Gradient Boosting Decision Tree (GBDT)
GBDT
TBD
XGBoost
Formula Derivation
Let’s talk about the
\[Obj = \sum_{i=1}^{N}l\left(y_i, \hat{y_i}\right) + \sum_{k=1}^{K} \cdot \Omega\left(f_k\right)\]objective functionfirst because every algorithm’s goal is to optimize the objective function. XGBoost algorithm’s objective function includes two parts: basic loss function $l$ and regularization term $\Omega$.
Here is the objective formula, where $N$ is the number of samples, $K$ is the number of boosting round/trees, $f(x_k)$ is the $k_{th}$ tree model in a function representation, $y_i$ is ground truth value of sample $i$, $\hat{y_i}$ is the predicted value of sample $i$.For a regression problem, the loss function can be $l(y_i, \hat{y_i}) = \frac{1}{2} \cdot \left(y_i - \hat{y_i}\right)^2$. For classification problem, it can be $l(y_i, \hat{y_i}) = - \left(y_i\cdot\log(\sigma(\hat{y_i})) + \right(1-y_i)\cdot\log(1-\sigma(\hat{y_i})))$
Next, let’s move to the
\[\hat{y_i} = \sum_{k=1}^{K}f_k\left(x_i\right)\]predicted value$\hat{y_i}$, just make sure know how $\hat{y_i}$ comes from.
In the boosting tree algorithm, the predicted value follows addictive model rule:If we now at $t_{th}$ boosting round say time $t$, the corresponding predicted values is $\hat{y_i}^{(t)} = \sum_{k=1}^{t}f_k(x_i)$, the $t_{th}$ tree model’s predicted is $f_t(x_i)$. Then, we have:
\[\hat{y_i}^{(t)} = \sum_{k=1}^{t}f_k(x_i) = \hat{y_i}^{(t-1)} + f_t(x_i)\]Ok, now we replace the $\hat{y_i}$ to
\[\begin{aligned} Obj^{(t)} &= \sum_{i=1}^{N}l(y_i, \hat{y_i}^{(t)}) + \sum_{k=1}^{t}\Omega(f_k)\\ &= \sum_{i=1}^{N}l(y_i, \hat{y_i}^{(t-1)} + f_t(x_i)) + \sum_{k=1}^{t}\Omega(f_k)\\ &= \sum_{i=1}^{N}l(y_i, \hat{y_i}^{(t-1)} + f_t(x_i)) + \Omega(f_t) + C \end{aligned}\]update objective functionat time $t$, notice that here we already know $t_{1:t-1}$ trees, so that $\sum_{k=1}^{t-1}\Omega(f_k)$ is a constant $C$.Then, how can we find a optimized $f_t$ to minimize the objective function at time $t$? Let’s apply some tricky meth on it. Recall the Taylor series expansions: $\begin{aligned} f(x+h)=& f(x)+h f^{\prime}(x)+\frac{h^{2}}{2} f^{\prime \prime}(x)+\cdots+\frac{h^{(n-1)}}{(n-1) !} f^{(n-1)}(x)+\frac{h^{n}}{n !} f^{n}(x+\lambda h) \end{aligned}$. We regard the $l(y_i, \hat{y_i}^{(t-1)})$ as $f(x)$ and $f_t(x_i)$ as $h$, take $n=2$. So, we have:
\[l(y_i, \hat{y_i}^{(t-1)} + f_t(x_i)) \approx l(y_i, \hat{y_i}^{(t-1)}) + [\partial_{\hat{y_i}^{(t-1)}}{l(y_i, \hat{y_i}^{(t-1)})}]\cdot f_t(x_i)+ [\frac{1}{2}\cdot\partial^{2}_{\hat{y_i}^{(t-1)}}{l(y_i, \hat{y_i}^{(t-1)})}]\cdot f_t^2(x_i)\]Let $g_i = [\partial_{\hat{y_i}^{(t-1)}}{l(y_i, \hat{y_i}^{(t-1)})}]$ and $h_i = [\partial^{2}_{\hat{y_i}^{(t-1)}}{l(y_i, \hat{y_i}^{(t-1)})}]$, note that $l(y_i, \hat{y_i}^{(t-1)})$ is constant cause for time $t-1$, $\hat{y_i}^{(t-1)}$ is already known. So, we have:
\[l(y_i, \hat{y_i}^{(t-1)} + f_t(x_i)) \approx l(y_i, \hat{y_i}^{(t-1)}) + g_i \cdot f_t(x_i) + \frac{1}{2}h_i \cdot f_t^2(x_i)\] \[\begin{aligned} Obj^{(t)} &\approx \sum_{i=1}^{N}[l(y_i, \hat{y_i}^{(t-1)}) + g_i \cdot f_t(x_i) + \frac{1}{2}h_i \cdot f_t^2(x_i)] + \Omega(f_t) + C\\ &\approx \sum_{i=1}^{N}[g_i \cdot f_t(x_i) + \frac{1}{2}h_i \cdot f_t^2(x_i)] + \Omega(f_t) \quad(\text{remove constant terms}) \end{aligned}\]Now we successfully separate $\hat{y_i}^{(t-1)}$ and $f_t(x_i)$, and the only variable in objective function at time $t$ is $f_t(x_i)$. But $\Omega(f_t)$ is still there and it is related to model structure, recall that $f_t$ is the tree model, we of course can find a way to connect $f_t(x_i)$ and $\Omega(f_t)$.
The next step is
\[f_{t}(x)=w_{q(x)}, \quad w \in \mathbf{R}^{T}, q: \mathbf{R}^{d} \rightarrow\{1,2, \cdots, T\}\]defining the tree modelin a mathematical way.
A tree model has $T$ leaf nodes, we can assign each leaf node a value, called ‘weight’. That means if the sample finally fall into that node, the tree model outputs that node’s weight value as output. Then, we have a weight vector $\omega$ with length of number of leaves for the tree model. $\omega_j$ means the weight of leaf node $j$.
We also define a mapping function that represents mapping the input sample to the tree model’s leaf node, say $q$. Input the sample $x$, $q(x)$ will output the leaf node index. Then, the $f_t(x)$ can be represented as follow:Next we
\[\Omega(f)=\gamma T+\frac{1}{2} \lambda\|w\|^{2} = \gamma T+\frac{1}{2}\lambda \sum_{j=1}^{T}\omega^2_j\]define the tree complexity$\Omega$, which is composed of number of tree leaf node and $L_2$ of leaf node weights.Each sample will finally fall into one leaf node, in another word, each leaf node has at least one sample in that node. We can use $I_{j}=\left{i q\left(x_{i}\right)=j\right}$ to represent the all samples that fall into node $j$. So, $f_t(x_i)$ means you input a sample $x_i$, then output a value. Which equals to $f_t(x_i) = \omega_{q(x_i)}$.
\[\begin{aligned} Obj^{(t)} &\approx \sum_{i=1}^{N}[g_i \cdot f_t(x_i) + \frac{1}{2}h_i \cdot f_t^2(x_i)] + \Omega(f_t)\\ &= \sum_{i=1}^{N}[g_i \cdot \omega_{q(x_i)} + \frac{1}{2}h_i \cdot \omega^2_{q(x_i)}] + \gamma T+\frac{1}{2}\lambda \sum_{j=1}^{T}\omega^2_j\\ &= \sum_{j=1}^{T}\bigg[(\sum_{i\in I_j}g_i )\omega_j + \frac{1}{2}(\sum_{i\in I_j}h_i)\omega^2_j\bigg] + \gamma T+\frac{1}{2}\lambda \sum_{j=1}^{T}\omega^2_j \qquad (\text{Use leaf node to represent it, samples with same node has same weight})\\ &= \sum_{j=1}^{T}\bigg[(\sum_{i\in I_j}g_i )\omega_j + \frac{1}{2}(\sum_{i\in I_j}h_i + \lambda)\omega^2_j\bigg] + \gamma T \end{aligned}\]
Finally, we have thefinal objective function with tree model definitions:Let $G_{j}=\sum_{i \in I_{j}} g_{i}$, $\quad H_{j}=\sum_{i \in I_{j}} h_{i}$ to simply, note that $G_j$ and $H_j$ are both constants.
\[Obj^{(t)} = \sum_{j=1}^{T}\bigg[G_j\omega_j + \frac{1}{2}(H_j+\lambda)\omega^2_j\bigg] + \gamma T\]Once we have the final objective function, we can easily find the
\[\hat{\omega}_j = -\frac{G_j}{H_j + \lambda} \qquad Obj = -\frac{1}{2}\sum_{j=1}^{T} \frac{G_j^2}{H_j + \lambda} + \gamma T\]optimized weight$\hat{w_j}$ for each leaf node $j$. If we fix the number of leafs $T$, then, we can compute the weight for each nodes (note that each node is independent) by applying quadratic formula:Finally, Let’s talk about how to decide $T$, because previous calculation we fix the $T$, now we have to handle it. I.e, how the tree grow? Similar to the decision tree grow, it use
\[\begin{aligned} Gain &= Obj_{L+R}-\left(O b j_{L} + Obj_{R}\right)\\ &=\left[-\frac{1}{2} \frac{\left(G_{L}+G_{R}\right)^{2}}{H_{L}+H_{R}+\lambda}+\gamma\right]-\left[-\frac{1}{2}\left(\frac{G_{L}^{2}}{H_{L}+\lambda}+\frac{G_{R}^{2}}{H_{R}+\lambda}\right)+2 \gamma\right]\\ &=\frac{1}{2}\left[\frac{G_{L}^{2}}{H_{L}+\lambda}+\frac{G_{R}^{2}}{H_{R}+\lambda}-\frac{\left(G_{L}+G_{R}\right)^{2}}{H_{L}+H_{R}+\lambda}\right]-\gamma \end{aligned}\]"Gain" to split tree nodes. Which means after that split, will objective function smaller? Use formula to represent it (note that the “gain” does not relate to $\omega$, it all depends on the first-order and second-order derivatives of loss function):Features
- Pre-sorting:
- For each node, enumerate over all features
- For each feature, sort the instances by feature value
- Use linear scan to decide the best split along that feature basis information gain.
- Take the best split solution along all the features
- Histogram-based:
- Split all data points for a feature into discrete bins
- Uses these bins to find the best split value of histogram
- Pre-sorting:
Feature importance
weight/frequency: the number of time a feature is used to split the data across all trees.
For example: feature#1 used 2, 3, 4 times to split nodes in tree#1, tree#2 and tree#3 respectively, feature#1 weight is $2+3+4=9$. Using this feature importance may have impact on binary features or features with less possible values, i.e. those features may have lower weight importance than features with mass possible values.The frequency for feature#1 is calculated as its percentage weight over weights of all features.
gain: The average information gain across all splits the feature is used in.
In summary:
- step 1: calculate previous node information entropy
- step 2: calculate all child nodes information entropy and get weighted sum
- step 3: differentia these two information entropy and weight it according the proportion of current samples affected by the split(in the example it is all samples)
- step 4: sum up across all trees and splits the feature is used in
cover: The average coverage across all splits the feature is used in
For example: feature#1 split 10, 5, 2 samples in tree#1, tree#2 and tree#3 respectively, feature#1 weight is $10+5+2=17$. This will be calculated for all features as a percentage for all features’ cover metrics.
LightGBM
- Gradient-based One-Side Sampling (GOSS)
- Keeps all the instances with large gradients and performs random sampling on the instances with small gradients
- Training instances with small gradients have smaller training error and it is already well-trained.
To achieve good balance between reducing the number of data instances and keeping the accuracy for learned decision trees, GOSS introduces a constant multiplier for the data instances with small gradients.
- Feature importance
- split: (identical to xgboost weight/frequency) The number of times the feature is used in the model
- gain: (identical to xgboost gain) Total gains of splits when feature is used
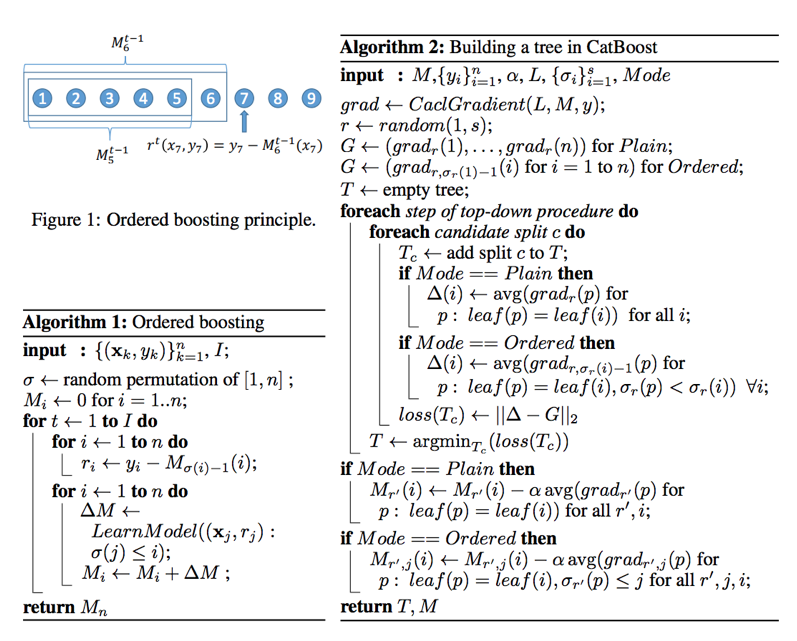
CatBoost
- Practical implement of orderd boosting
- Permutating the set of input observations in a random order, multiple random permutations are generated
- Converting the label value from a floating point or category to an integer
- Transform all categorical feature to numeric values using avg_target=(countInClass+prior)/(totalCount+1)
- countInClass is how many times the label value was equal to “1” for objects with the current categorical feature value.
- TotalCount is the total number of objects that have a categorical feature value matching the current one
- Feature importance
- predicted value changes: How munch on average the prediction changes if the feature value changes. The bigger the value of the importance, the bigger on average is the change to the prediction value.
- loss function change: Represents the difference between the loss value of the model with this feature and without it. The model without the feature is equivalent to the one that would have been trained if this feature was excluded from the dataset. Since it is computationally expensive to retrain the model without one of the features, this model is built approximately using the original model with this feature removed from all the trees in the ensemble.
- internal feature importance: Importance of values both for each of the input features and their combinations.
- prediction difference: Impact of a feature on the prediction results for a pair of objects. For each feature PredictionDiff reflects the maximum possible change in the predictions difference if the values of the feature is changed for both objects. The change is considered only if it affects the ranking order of objects.